Contents
Gene Technology
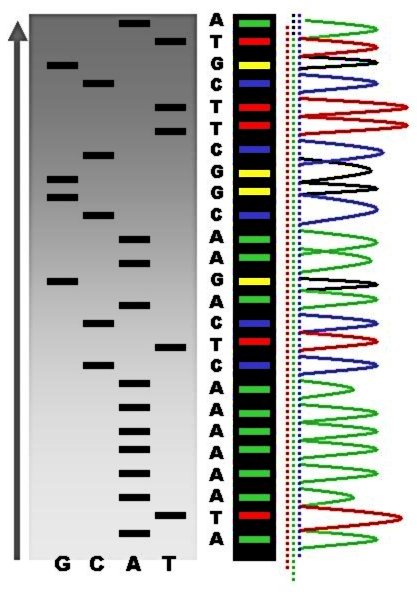
Sequencing Projects
Sequencing projects have read the genomes of a wide range of organisms, including humans, which provides opportunities to screen DNA to identify potential medical problems. The human genome consists of 3 billion base pairs and approximately 20,000 genes. The proteome is all the proteins in a cell and this varies over time and between cells.
With simpler organisms, if the genome sequence is known then the proteome can be derived from the genetic code. This may have many applications, including the identification of potential antigens for use in vaccine production. In more complex organisms like humans, the presence of non-coding DNA and regulatory genes means that knowledge of the genome cannot easily be translated into the proteome. Sequencing methods are continuously updated and have become automated.
Recombinant DNA Technology
This technology involves the combining of different organisms' DNA, which could enable scientists to manipulate and alter genes to improve industrial processes and medical treatment. Transferring DNA fragments between/within species is possible because the genetic code is universal, and transcription and translation work the same in all organisms too.
The first step in this technology is to produce or isolate the fragment of DNA to be recombined with another piece of DNA. There are three methods to create fragments of DNA:
Reverse Transcriptase
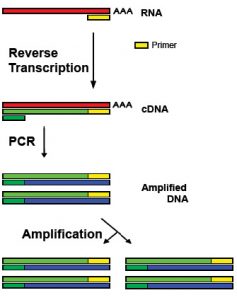
The enzyme reverse transcriptase can make DNA copies from mRNA, and this naturally occurs in certain viruses, such as HIV. A cell that naturally produces the protein that scientists want to harvest is selected. These cells should have large amounts of mRNA for the protein. The reverse transcriptase enzyme can then align and join the DNA complementary bases that match the mRNA sequence. This is single-stranded and known as complementary DNA (cDNA). To make this DNA fragment double-stranded, the enzyme DNA polymerase is used. The cDNA is intron-free because it is based on the mRNA template.
Restriction Endonucleases
Restriction endonucleases are enzymes that cut up DNA, which naturally occur in bacteria as a defense mechanism. There are many restriction enzymes that have an active site complementary in shape to a range of different DNA base sequences, described as recognition sequences, and therefore each enzyme cuts the DNA at a specific location. Some enzymes cut at the same location in the double strand and create a blunt end, while other enzymes cut to create staggered ends and exposed DNA bases. The exposed staggered ends are palindromic and referred to as 'sticky ends' because they have the ability to join to DNA with complementary base pairs.
Gene Machine
DNA fragments can be created in a lab using a computerized machine. Scientists first examine the protein of interest to identify the amino acid sequence, and from that work out what the mRNA and DNA sequence would be. The DNA sequence is then entered into the computer, which checks for biosafety and biosecurity to ensure that the DNA being created is safe and ethical to produce. The computer can create small sections of overlapping single strands of nucleotides that make up the gene, called oligonucleotides. The oligonucleotides can then be joined to create the DNA for the entire gene. PCR can be used to amplify the quantity and to make the double strand. This process is very quick, accurate, and makes intron-free DNA so it can be transcribed in prokaryotic cells.
Amplifying DNA Fragments
Once the DNA fragment has been isolated, it needs to be cloned to create large quantities of it. This can be done in vivo or in vitro.
In Vitro
Fragments of DNA can be amplified in vitro via the polymerase chain reaction (PCR). This is done using an automated machine. The temperature is first increased to split the DNA into single strands, and then cooled so that a primer can attach. The enzyme DNA polymerase then attaches complementary free nucleotides and makes a new strand to align next to each template.
In Vivo
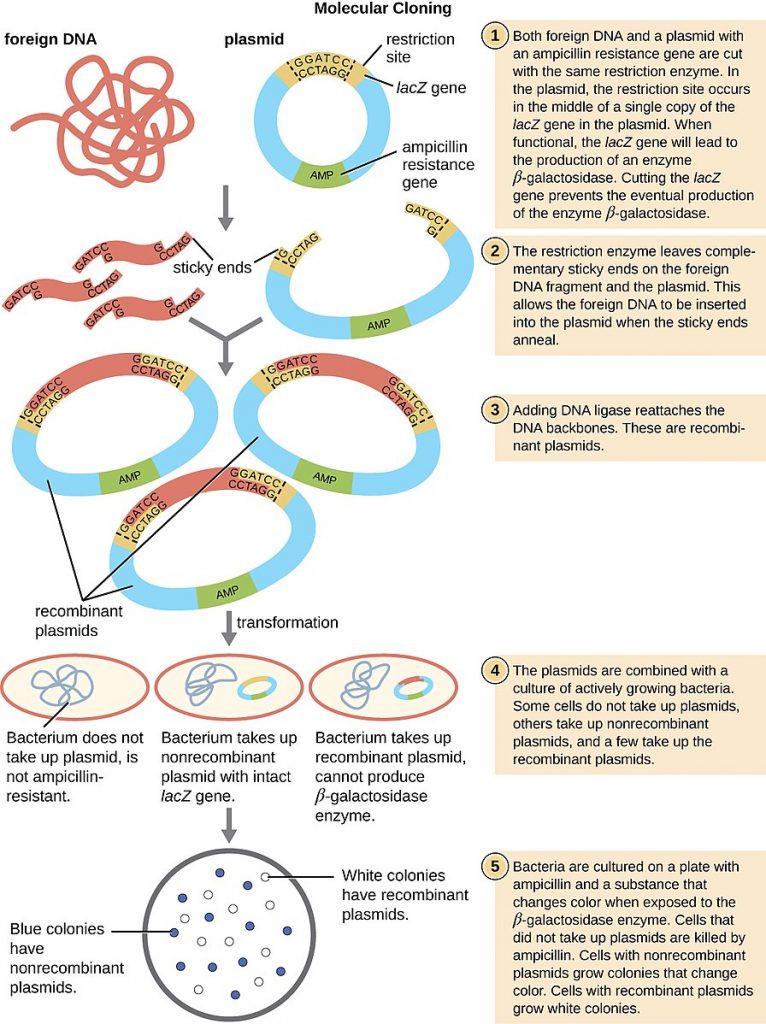
Firstly, restriction endonucleases are used to cut DNA, leaving sticky ends. The DNA fragment then needs extra DNA added to enable RNA polymerase to attach and transcribe mRNA from it. The DNA that is added is called a promoter and a terminator region, to enable transcription to be initiated and to finish. This DNA then needs to be joined to a vector, which will transport the DNA into the host cell. Plasmids are commonly used vectors. Restriction endonucleases are used to cut open the plasmid, the bacterial loop of DNA. Using the same restriction endonuclease to create the DNA fragment and to cut open the plasmid ensures they have complementary sticky ends. DNA ligase is used to stick the DNA fragment in. This is known as recombinant DNA now.
The recombinant plasmids are then introduced into the bacteria by transformation. Transformation involves mixing the plasmids and bacterial cells in a solution of calcium ions. Sudden changes in the temperature of the solution make the bacterial membrane more permeable and the plasmid enters. Not all the plasmids will enter the bacterial cell, and not all the plasmids will have taken up the DNA fragment, so the next step is to identify which bacterial cells did take up recombinant plasmids.
Plasmids contain antibiotic resistance genes. To identify if the plasmid was taken up, the bacteria are grown on agar plates containing antibiotics, such as ampicillin. If the bacteria took up the plasmid, then they will have the gene for ampicillin resistance and will survive. This is the first step of identification, as we know these bacteria contain a plasmid.
Next, further testing is needed to identify if the plasmid they took up was a recombinant one. The DNA fragment is deliberately inserted in the middle of a marker gene, which could be a gene for resistance to another antibiotic or a fluorescent protein gene. By inserting the DNA fragment in the middle of a marker gene, that protein will no longer be made, so the bacteria won't be resistant to another antibiotic or it will not produce a fluorescent protein. Out of the cells that survived in the ampicillin, these can be tested further. Those that do fluoresce under UV light must contain a non-recombinant plasmid, but those that do not fluoresce must have the recombinant plasmid. In this way, the bacteria containing the recombinant DNA has been identified. These bacteria can be grown, and the DNA fragment is therefore amplified.
DNA Probes
DNA probes are short, single-stranded pieces of DNA that are labeled radioactively or fluorescently so they can be identified. This is used to locate specific alleles of genes and to screen patients for heritable conditions, drug responses, or health risks. A sample of patient DNA is removed and heated to make it single-stranded. It is mixed with DNA probes that have been created to be complementary to a range of different alleles for genes (e.g., for different diseases). If the patient has that allele, their DNA will bind with the probe, and this can be identified using an x-ray or UV light.
Genetic Fingerprinting
95% of human DNA is made up of introns, which consist of many variable number tandem repeats (VNTRs). The probability of two individuals having the same VNTRs is very low, however, the more closely related you are, the more similar the VNTRs are.
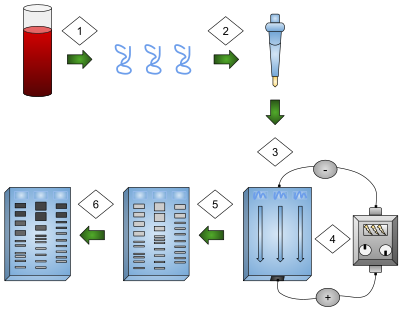
The technique of genetic fingerprinting is the analysis of VNTR DNA fragments that have been cloned by PCR, and its use in determining genetic relationships and in determining the genetic variability within a population.
This method can be used in the fields of forensic science to place suspects at crime scenes, for medical diagnosis, and to ensure animals and plants are not closely related before being bred.
- Why is PCR often used when producing genetic fingerprints?
- amplify
- How are sticky ends created?
- Your answer should include: restriction / endonucleases
- What enzymes are used to create cDNA?
- Your answer should include: reverse / transcriptase
- What is the commonly used vector in in vivo gene cloning?
- plasmid
- What is a DNA probe?
- Your answer should include: short / single / stranded / piece / labeled / DNA
- Which sections of the DNA are examined in genetic fingerprinting?
- VNTRs